以哈克尼西棉细胞质陆地棉雄性不育系及其保持系、恢复系为材料, 克隆并测序线粒体功能基因的侧翼序列。通过生物信息学分析, 在
The cytoplasmic male sterile (CMS) line with the cytoplasm of
线粒体编码的蛋白质主要是氧化呼吸链的结构蛋白和生物合成蛋白, 在能量代谢中起关键作用[1,2]。植物线粒体影响一些重要的农艺性状, 还参与细胞质雄性不育(cytoplasmic male sterility, CMS)形成与遗传。植物细胞质雄性不育在高等植物中普遍存在, 早前在43个科162个属320种植物中发现617例天然或种属间杂交来源的雄性不育[2,3]。CMS与线粒体DNA的重组重排和一些基因的特异性表达有关[4]。相关研究在杂种优势利用和核质互作分析方面具有重要的意义。
植物的线粒体基因组非常复杂[5,6], 包括内含子的顺式和反式剪切[7], 重复序列的重组重排[8], 外源序列的迁入[9], 亚分子计量序列的变异[10], RNA加工[11], RNA编辑, 转录后调控[12,13,14], 细胞核基因的调控等[15,16,17,18]。这些伴随进化而不断发生的变异, 是引起细胞质雄性不育的重要原因[6]。
迄今, 已在许多植物中相继发现CMS基因并阐释其败育机制, 例如玉米的CMS-T ( urf13/ atp4)[19], CMS-S ( orf355+ orf77/ atp9)[20], 芸薹属CMS-nap ( orf222/ nad5c/ orf139)[21], CMS-pol ( orf224/ atp6)[22], CMS-ogrua ( orf138/ atp8)[23], CMS-junce a ( orf108/ atpA)[24], 小麦CMS-T ( orf256/ cox1)[25], 向日葵CMS-pet1 ( orf522/ atpA)[26], 水稻CMS-BT ( orf79/ atp6)[27], CMS-HL ( orfH79/ atp6)[28], 萝卜CMS-kos ( orf125/ atp8)[29], 以及矮牵牛CMS-S ( pcf/ nad3/ rps12)[30]。这些orf具有胞质和种属特异性, 多由线粒体保守序列与未知序列组成[31]; 与功能基因共转录, 影响线粒体中ATP合成途径, 造成能量供给不足, 致使花器官不能正常行使功能, 导致败育。
棉花是重要的天然纤维作物, 同时也是重要的油料作物, 其主要栽培种陆地棉( Gossypium hirsutumL.)具有显著的杂种优势[32]。棉花细胞质雄性不育研究始于20世纪60年代, 通过远缘杂交和回交获得以陆地棉核为背景的分别具有亚洲棉( G. arboreumL.)、异常棉( G. anomalumWawr. & Peyr)和哈克尼西棉( G. harknessiiBrand.)细胞质的胞质雄性不育系, 其中哈克尼西棉细胞质不育系的不育性遗传稳定。1988年我国实现了哈克尼西棉胞质CMS三系配套, 稍后的陆地棉三系、海岛棉三系配套研究, 为陆地棉三系育种提供了丰富的基础材料; 棉花三系广泛应用于生产实践中, 但是棉花的细胞质雄性不育、育性恢复以及杂种优势等的分子遗传机理仍不清楚[33,34]。现有研究主要集中于线粒体基因组结构、转录和翻译产物等方面的差异, 然而结果不尽相同。采用RAPD技术分析棉花CMS三系线粒体基因组和叶绿体基因组的差异发现, CMS主要与线粒体异常有关。以哈克尼西棉CMS系和保持系的花药和黄化苗为材料, 分别对线粒体蛋白质进行SDS-PAGE、RAPD和RFLP分析比较, 不育系与保持系线粒体相比, 发现缺少一个大小为1.9 kb与 cox2基因具有同源序列的片段, 表达产物缺少约31 kD的多肽[35]。通过RFLP分子标记技术, 发现 cox2探针在陆地棉不育系CMS-D2-2比保持系多出1条片段[36]。以哈克尼西棉CMS系和保持系为材料, 得到扩增片段大小存在差异的 atpA基因及其侧翼序列; RACE方法获得 atpA基因cDNA全长测序后发现, 不育系为1582 bp, 保持性为1566 bp, 两者预测的编码蛋白质不同, 认为这可能是导致CMS的原因[37,38]。
本研究通过比较基因组方法, 克隆一个位于 atp4下游的差异orf160, 构建表达载体, 进行转基因验证, 通过转化酵母, 发现 orf160影响酵母正常生长; 在拟南芥和烟草中, orf160的表达可能影响转基因后代花粉的形成, 导致后代结实率和种子活力下降, 可能在导致不育方面发挥一定的作用。
陆地棉不育系2074A遗传稳定, 是哈克尼西棉细胞质CMS-D2-2不育系, 由原始不育系DES-HAMS 277经过多代回交选育而来, 系谱为({[(DES-HAMS 277×E369)×中7] BC13F1×鄂棉18}BC12F1×徐244) BC6F1。保持系2074B是苏棉20 (徐棉244), 恢复系E5903是Z834R, 来自DES-HAF 277的多代选育后代[33]。
哥伦比亚野生型拟南芥( Arabidopsis thaliana columbia)和本氏烟草( Nicotiana benthamiana)用于转化。大肠杆菌( Escherichia coli) DH5α, 根瘤农杆菌( Agrobacterium tumefaciens) GV3101, 植物表达载体pCAMBIA2301M和pCF203、酵母表达载体p426HXT7均由本实验室保存。
棉花线粒体DNA/RNA提取, 包括线粒体分离和线粒体裂解2个主要环节[39]。
将脱绒的棉花种子种植于经过灭菌处理的沙土, 28℃暗培养4~6 d; 取200~300 g黄化苗在匀浆缓冲液(300 mmol L-1蔗糖, 20 mmol L-1 KH2PO4, 10 mmol L-1 KCl, 5 mmol L-1 EDTA-Na2, 10 mmol L-1 Tris-HCl, 0.1% ( W/V) BSA, 6 mmol L-1 β-mercaptoethanol, pH 7.5)中差速匀浆, 6层纱布过滤, 低速离心去除细胞核和细胞碎片; 用漂洗缓冲液(300 mmol L-1蔗糖, 20 mmol L-1 KH2PO4, 10 mmol L-1 KCl, 5 mmol L-1 EDTA-Na2, 10 mmol L-1 Tris-HCl, 0.1% ( W/V) BSA)漂洗2次, 去除大分子杂质; 获得粗制线粒体, 加入悬浮缓冲液用毛刷将其轻轻刷起,转到蔗糖密度梯度(蔗糖梯度18%、36%、52%)上, 40 000× g离心90 min, 吸取36%~52%之间的线粒体, 用漂洗缓冲液漂洗2次, 即为完整的线粒体。
用CTAB裂解液(0.1 mol L-1 Tris-HCl, 0.02 mol L-1 EDTA-Na2, 2% CTAB, 1.4 mol L-1 NaCl, 2% PVP 40, 2%~3% β-疏基乙醇)裂解线粒体, 无水乙醇沉淀DNA, 70%乙醇漂洗2次, 得到纯度较高、较完整的线粒体DNA。
用DEPC处理过的CTAB裂解液裂解线粒体, 无水乙醇沉淀RNA, 得到纯度较高、较完整的线粒体RNA (华金平等, 中国发明专利, 申请号2010 10268760.2), 利用TaKaRa试剂盒, 随机引物反转录获得线粒体cDNA。
利用生物信息学比较不育系、保持系和恢复系线粒体功能基因的侧翼序列, 获得差异基因 orf, 设计相应引物, 扩增全长并测序验证。利用对应的orf引物, 以RT-PCR检测差异 orf在“三系”中的表达情况, 最终确定差异 orf160。
以溶菌酶法获得酵母Y189的DNA, 扩增获得 cox4的序列[40,41]。连接至pMD18-T easy vector, 挑选单克隆测序, 选择正向插入的单克隆。利用 SalI和 Hind III双酶切pMD18- cox4质粒DNA和 orf160, T4连接, 用引物cox4F和orf160R2组合扩增 cox4和 orf160, 将酶切位点替换为 BamH I和 SacI, 并用这2个限制性内切酶对PCR产物、pCAMBIA2301M和pCF203质粒DNA进行双酶切, T4连接, 转化大肠杆菌感受态DH5α, 挑取阳性克隆进行验证, 即完成含有线粒体导肽( cox4)的植物表达载体和GFP瞬时表达载体的构建, 重组质粒分别被命名为pCAM BIA2301M- cox4- orf160和pCF203- cox4- orf160。将构建好的植物表达载体和瞬时表达载体提取质粒, 转化农杆菌感受态GV3101, 用于遗传转化。利用农杆菌介导法将植物表达载体转化拟南芥和烟草。利用基因枪法将瞬时表达载体转化洋葱表皮细胞, 观察线粒体导肽定位情况。
酵母表达载体构建: 在pMD18-T easy vector上直接用 BamH I和 Hind III双酶切质粒DNA, 将回收片段与p426HXT7连接, 即为p426HXT7- cox4- orf160; 试验处理还包括阳性对照(p426HXT7- GhPAT1), 插入基因 GhPAT1和阴性对照(p426HXT7)。转化试验3次重复, 分别涂于尿嘧啶缺失抗性平板, 观察酵母生长情况。
阳性植株的鉴定, 通过2轮4次筛选。首先在50 mmol L-1卡那霉素MS平板上筛选抗性绿苗, 移栽绿苗; 三叶期取叶片提取总DNA, 用卡那霉素抗性基因标记Kan、线粒体导肽cox4标记和基因 orf160标记引物扩增。
| 表1 本研究使用的引物序列Table 1 Oligonucleotides used in this study |
拟南芥T1和T2代种子, 用经5%的次氯酸钠溶液(0.1%的Triton X-100)表面消毒15 min和灭菌水洗涤; 铺种在50 mmol L-1卡那霉素的MS培养基上, 4℃春化处理2 d后, 转至21℃、光照16 h/黑暗8 h和60%~75%相对湿度的转基因室生长一周后, 统计分离比。选择绿苗移栽到无菌土中(蛭石和营养土, 体积比2∶1); 单株收获T1和T2代植株, 统计结 实率。
用5%的次氯酸钠溶液(0.1%的TritonX-100)表面消毒烟草T1和T2代种子20~30 min, 用灭菌水洗涤后铺种含50 mmol L-1卡那霉素的MS培养基, 在28℃、光照16 h/黑暗8 h和60%~75%相对湿度的条件下培养1周, 调查T2代烟草发芽率。烟草苗直接在28℃、光照16 h/黑暗8 h和60%~75%相对湿度的无菌室中培养。烟草T2代植株生长缓慢, 且长势弱。取开花当天的烟草T1代和拟南芥T1、T2代植株花粉, 利用I2-KI染色, 观察败育情况。单株收获, 统计结实率。
将转化后的酵母涂在尿氨酸缺失的YPD (yeast extract peptone dextrose)培养基上, 30℃培养2~3 d, 观察酵母生长情况, 分析 orf160表达产物对酵母生长的影响。
orf160上游为功能基因 atp4。设计不同引物PCR验证, 在保持系和恢复系中没有获取扩增条带(图1), 证明 orf160在保持系和恢复系中不存在。
在不育系中扩增获得 orf160全长480 bp, N端与 atp6编码序列同源, C端含有部分序列与核编码序列同源, 编码159个氨基酸; 氨基酸序列与链霉菌属蛋白序列具有一定的同源性, 与紫黑链霉菌膜蛋白[putative membrane protein, gi|345007990 ( Streptomyces violaceusniger Tu 4113)]和灰黄链霉菌细胞周期蛋白(cell cycle protein, gi|302559642 ( Streptomyces griseoflavus Tu4000))同源性分别为28%与31% (图2)。
 | 图1 orf160基因的克隆不同mtDNA模板和3对orf160引物的PCR扩增结果。图中, mtDNA模板依次是: 1, 4, 7: 保持系2074B; 2, 5, 8: 恢复系E5903; 3, 6, 9: 不育系2074A。M: D2000。样品1, 2, 3的扩增引物为orf160F3与orf160R3; 样品4, 5, 6的扩增引物为orf160F4与orf160R4; 样品7, 8, 9的扩增引物为orf160F5与orf160R5。Fig. 1 Cloning of orf160PCR amplified products of mtDNA in different lines using different primers of orf160. 1, 4, 7: maintainer line 2074B; 2, 5, 8: restore line E5903; 3, 6, 9: CMS line 2074A. Primers used in amplification as follow: orf160F3 and orf160R3 for the sample 1, 2, 3; orf160F4 and orf160R4 for the sample 4, 5, 6; orf160F5 and orf160R5 for the sample 7, 8, 9. |
 | 图2 orf160基因序列分析ORF160蛋白与其他相关蛋白的氨基酸序列比对分析。序列比对图中, 完全相同与基本相同的氨基酸残基分别用黑色与灰色背景表示。Fig. 2 Sequence analysis of orf160Amino acid sequence alignment of ORF160 with other related proteins. Identical and conservative residues are denoted by black shading, grey shading, respectively. |
以酵母DNA为模板, 扩增获得线粒体导肽78 bp, 引物参见Kim等[41], 插入到pMD18-T easy vector, 单克隆测序; 选择正向插入 cox4片段的单克隆, 提取质粒DNA, 酶切回收获得 cox4片段; 另外从不育系线粒体DNA中扩增 orf160, 酶切回收, 分别将 cox4和 orf160插入到载体pCAMBIA2301M, 重组质粒被命名为pCAMBIA2301M- cox4- orf160(图3-A)。同时, 构建了GFP (green fluorescent protein)瞬时表达载体, 将 cox4和 orf160片段插入到含有GFP基因的载体pCF203中, 重组质粒被命名为pCF203- cox4- orf160(图3-B), 转化洋葱表皮细胞, 确定 cox4导肽的作用。在荧光电子显微镜下, 只有细胞膜上观察到绿色荧光, 而细胞核中没有观察到绿色荧光, GFP只被定位到细胞膜的周围(图4-A); 而阳性对照即空载质粒转化洋葱表皮细胞, 细胞膜和细胞核中都能观察到绿色荧光, GFP被定位到细胞核和细胞膜上(图4-B)。
 | 图3 植物表达载体的构建A: 拟南芥和本氏烟草表达载体pCAMBIA2301M- cox4- orf160; B: 瞬时表达载体pCF203 -cox4- orf160。Fig. 3 Vector of pCAMBIA2301M- cox4- orf160 and pCF203- cox4- orf160 for plant transformationA and B: Constructed vectors of cox4and orf160 for Arabidopsis and Nicotiana Benthamiana transformation. The diagram shows the strategy for constructing each cloned gene into the modified pCAMBIA2301M and pCF203 plant transformation vector. The GFP indicates the green fluorescent protein (GFP). NPTII: Neomycin phosphotransferase II gene; MCS: multiple cloning site; NOS ter: NOS terminator site; LB: left border; RB: right border; Kmr: kanamycin resistance gene. |
 | 图4 GFP瞬时表达载体转化洋葱表皮细胞的结果A: 瞬时表达载体荧光显微镜下镜检; B: 瞬时表达载体阳性对照。Fig. 4 Results of transformation of onion epidermal cell with GFP transient expression vectorA: Image of pCF203- cox4- orf160 GFP fluorescence in the onion transient expression assay; B: Image of pCF203 GFP fluorescence in the onion transient expression assay. |
构建酵母表达载体, 以p426HXT7为载体, 插入 cox4和 orf160, 即为p426HXT7- cox4- orf160; 同时将阳性对照p426HXT7插入棉花功能基因 GhPAT1 (载体p426HXT7- GhPAT1)(图5和图6), 转化后涂在尿氨酸缺失的YPD培养基上, 30℃培养2~3 d, 观察到空白对照和阳性对照材料酵母转化正常, 而p426HXT7- cox4- orf160转化后酵母生长异常, 酵母斑较少(图7)。说明 orf160的表达对酵母的生长有一定抑制作用。
将拟南芥T1植株铺种在50 mmol L-1卡那霉 素抗性的MS平板上, 筛选抗性绿苗(图8-A); 提取移栽绿苗总DNA, 利用卡那霉素抗性标记(NPTII) Kan进行PCR检测(图9), 从T1代植株筛选得到3棵阳性植株, 编号为T101、T102和T103, 其中T103植株长势较好, T101和T102生长较慢, 结实率低。
 | 图5 酵母表达载体构建A: 含有 cox4- orf160酵母表达载体p426HXT7- cox4- orf160构建图; B: 含有 GhPAT1阳性对照载体p426HXT7- GhPAT1构建图。Fig. 5 Vector construction of p426HXT7- cox4- orf160 and p426HXT7- GhPAT1A: Vector construction of p426HXT7- cox4- orf160; B: Vector construction of p426HXT7- GhPAT1as a control. |
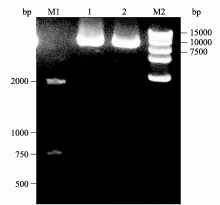 | 图6 酵母表达载体构建酶切验证限制性内切酶 BamH I和 Hind III双酶切验证载体构建, 1和2阳性克隆质粒DNA经过 BamH I和 Hind III双酶切电泳检测结果; M1: marker D2000; M2: marker D15000。Fig. 6 Verification of vector construction for yeast transforming by restriction endonuclease analysisRestriction analysis of p426HXT7- cox4- orf160using BamH I and Hind III two restriction endonuclease; M1: marker D2000; M2: marker D15000. |
T2代植株黄绿苗符合1∶3分离比(原始数 据: T101为12∶32, T102为29∶32, T103为 25∶93)。在电子显微镜下观察T103的T2代植株花器官, 花粉少量散出(图8-B), 花粉粒着色浅(图8-C); 野生型本氏烟草花粉粒饱满, 且染色较深(图8-D)。
对转基因烟草T1代植株, 首先进行PCR检测, 筛选出阳性植株(图10), 收获17个阳性单株, 依次编号为T01-T17。然后, 对T1代植株T01单株的花粉进行I2-KI染色, 发现花粉粒大小不均一, 形状不同, 着色深浅也不同, 说明花粉粒活性不相同, 畸形的花粉粒可能无活性或活性较低(图11-a); 而野生型烟草的花粉粒着色较深(图11-d)。观察种子发现, T1代转基因烟草种子多数不饱满、畸形干瘪(图11-b, e)。种子萌发实验表明, T1代种子发芽率为67.3% (图11-c), 野生型烟草的种子发芽率为99.7% (图11-f), 说明 orf160的表达影响转基因烟草后代的种子 发育。
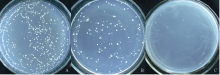 | 图7 酵母表达载体转化子检测A: 空白对照; 空载体p426HXT7转化酵母; B: 阳性对照; p426HXT7- GhPAT1转化酵母; C: p426HXT7- cox4- orf160转化酵母。Fig. 7 Results of the yeast transformantsA: Yeast colonies transformed by vector p426HXT7; B: yeast colonies transformed by vector p426HXT7- GhPAT1; C: Yeast colonies transformed by vector p426HXT7- cox4- orf160. |
 | 图8 转基因拟南芥生物学表型观察A: T1代转基因拟南芥抗性平板上筛选阳性植株; B: T2代转基因拟南芥花器官; C: T2代转基因拟南芥花粉粒I2-KI染色(×100);D: 野生型拟南芥花粉粒I2-KI染色(×100)。Fig. 8 Biological phenotypes of transgenic ArabidopsisA: transgenic Arabidopsis plants of T1 generation on resistance flat-panel; B: floral organs of T2 generation transgenic Arabidopsis;C: I2-KI stained pollen grains of T2 generation transgenic Arabidopsis; D: I2-KI stained pollen grains of wild Arabidopsis. |
 | 图9 转 cox4- orf160基因拟南芥Kan标记PCR检测结果M: marker D2000; A: T1代阳性单株; B: T2代单株。Fig. 9 PCR result of transgenic cox4- orf160 Arabidopsis plants with primer of Kan resistance markM: marker D2000; A: monoclonal individuals from T1 generation; B: monoclonal individuals from T2 generation. |
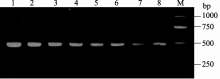 | 图10 转基因烟草T1代阳性株Kan标记PCR检测结果M: marker D2000; 1~8: T1代阳性单株。 M: marker D2000; 1-8: T1 monoclonal individuals.Fig. 10 PCR result of transgenic cox4- orf160 tobacco lines with primer of Kan resistance mark |
调查10个转基因烟草单株结实率、株高和第一果枝高度(图12), 发现野生型植株和转基因植株第一果枝高度( P=0.02)和第一果节位( P=0.035), 存在显著差异, 其余性状如株高、果枝数和种子总数无显著差异, 种子总数虽然未达显著差异, 但是总体要比野生型的数量少, 而且有一株没有结实, 说明转基因 orf160对种子发育和节间伸长有一定的影响。
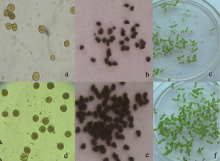 | 图11 转基因烟草T1代生物学表型观察a: T1代转基因烟草花粉粒I2-KI染色; d: 野生型花粉粒I2-KI染色; b: T1代转基因烟草种子; e: 野生型烟草种子; c: T1代转基因烟草种子发芽试验; f: 野生型烟草种子发芽试验。Fig. 11 Biological phenotypes of T1 generation transgenic tobaccoa: I2-KI stained pollen grains of T1 generation transgenic tobacco; d: I2-KI stained pollen grains of wild type tobacco; b: seeds of T1 generation transgenic tobacco; e: seeds of wild type tobacco; c: seeds germination test of T1 transgenic tobacco; f: seed germination test of wild type tobacco. |
种植拟南芥T1代3个单株全部种子, 随机选择T2代单株移栽。PCR鉴定的阳性植株119个, 其中正常植株32株, PCR鉴定的阴性植株的表型与野生型相同, 完全可育; 部分不育株73株, 完全不育株14株, 即约70%的植株表现部分不育或完全不育(表2)。
 | 图12 转基因烟草T1代植株和野生型植株生物学性状比较A: 转基因与野生型烟草T1代植株株高, 第一果枝高度和果枝数; B: 转基因与野生型烟草种子数。Fig. 12 Comparison of biological characters on individuals of T1transgenic tobacco and wild type tobaccoA: plant height, the first branch height and fruit branch number of transgenic T1 generation plant and wild type tobacco;B: total seed number of transgenic and wild type tobacco. |
转基因烟草T1代12个单株, 种植每个单株T2代的6个单株, 共72个单株。PCR随机鉴定、移栽的阳性单株37个, 25个植株表现部分不育或完全不育(表2)。
在调查拟南芥转基因植株中, T3代没有发现明显的雄性不育共分离的现象, 分离比差异大(表3)。对于T0118植株, T3代植株没有黄苗, 全是绿苗; T0128、T0215、T0369和T0360的T3代植株后代也多是绿苗。
卡那霉素基因作为选择标记基因之一, 应用 于转基因实验的大田试验植株活体表型鉴定和室 内阳性植株鉴定, 方便快捷。pCAMBIA2301M载 体中T-DNA内的 NPTII是新霉素磷酸转移酶基 因, 编码蛋白具有卡那霉素抗性, 用于转基因植 株的筛选; T-DNA外的Kmr也是编码卡那霉素抗 性基因, 用于载体构建过程中阳性单菌落筛选。而用于PCR筛选的Kan标记是根据这个基因的保守 序列设计的, 在阳性单克隆和阳性植株中都能被检测到。
| 表2 转 orf160基因拟南芥和烟草后代育性Table 2 Male sterility of transgenic orf160 plants from Arabidopsis and tobacco |
| 表3 转基因 orf160拟南芥转基因T2代分离Table 3 Segregation of the orf160 in Arabidopsis T2 transgenic plants |
未知功能orf存在部分同源序列。这种同源性与线粒体基因组、核基因组进化是否相关, 目前还不清楚。 orf160的N端与线粒体功能基因 atp6序列部分同源, C端与核序列部分同源。其他作物不育基因结构已有类似的报道, 如水稻BT- orf79和HL- orfH79 N端与 cox1序列相似, C端与微球菌 Kocuria rhizophila DC2201的putative ABC transporter permease/ATP-binding protein相似[27,28]。一般来说, 动物线粒体基因组比较保守, 而植物基因组变异广泛[5,6]。在进化过程中, 植物线粒体基因组经历了多种多样的基因组变异, 其中包括来自叶绿体和/或核基因组广泛的基因转移, 发生一系列的重组、重排、重复等变化, 产生许多未知orf。
线粒体转录组非常复杂, 功能基因表达不尽相同, 最重要的是未知功能orf的表达各异[42,43]。目前对线粒体转录组测序的相关报道不多。从转录水平上推测, 这些未知的orf表达差异很大, 而细胞质雄性不育通常就是未知功能的orf差异表达造成的。本研究通过扩增获得不育系和保持系线粒体功能基因的的侧翼序列, 在不育系中 atp4下游发现一个全长480 bp特有的 orf160, 功能同样未知。 cox4是克隆于酵母的基因, 仅78 bp, 利用 cox4作为线粒体导肽靶向定位应用广泛, 如向日葵不育基因 orf522和辣椒不育基因 orf456等线粒体转基因载体中[41,44]。本实验瞬时表达载体亚细胞定位结果显示, 外源 orf160确实能在线粒体中正常表达, 这与Kim等[41]报道结果相同, 即 cox4能定位到线粒体中。
前人报道, 败育orf在不同的植物种中不一定能够导致败育, 例如萝卜和油菜 Ogura-orf138转入拟南芥没有产生不育表型。所以, 本研究将 orf160转基因导入拟南芥和烟草中, 转基因植株没有产生完全的败育, 并不能够直接排除 orf160是哈克尼西棉细胞质的不育基因。事实上, 转基因后代均表现出部分不育; 拟南芥T2代植株不育和可育以3∶1的分离比分离(表3); 转基因植株结实率、种子发芽势、花粉生活力降低; 转基因烟草的花粉着色浅、种子出现明显畸形, 说明 orf160表达影响转基因受体的正常发育; orf160对酵母的生长的抑制作用也进一步证明了这一点, 相关机理以及机制、编码蛋白是否具有毒性还需进一步实验验证。
不育系 atp4下游发现的 orf160, 全长480 bp, 编码159个氨基酸序列与膜蛋白和细胞周期蛋白具有部分同源性, 该基因的转基因酵母菌斑畸形、生长缓慢; 转基因拟南芥和烟草后代植株的结实率降低, 种子大部分畸形, 且发芽率低。说明 orf160基因的表达影响转基因受体的正常发育。
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
|

