第一作者联系方式: E-mail: wangjia0724@126.com **同等贡献(Contributed equally to this work)
株高、分枝数及第1分枝高是油菜重要的农艺性状。本研究利用甘蓝型油菜GH06和P174杂交, F2通过单粒法连续自交至F11构建重组自交系群体, 利用油菜60K芯片对该群体进行基因分型, 构建高密度遗传连锁图谱。结果表明, 该图谱包含2795个SNP多态性标记位点, 总长1832.9 cM, 相邻标记间平均距离为0.66 cM。在此图谱基础上采用复合区间作图法(CIM), 检测到3个农艺性状的24个QTL。其中11个株高QTL分别位于A01、A06、A07、A08、A10和C06染色体, 单个QTL解释5.00%~15.26%的表型变异; 7个第1分枝高QTL分别位于A06、C05和C06染色体, 单个QTL解释5.04%~12.99%的表型变异; 6个分枝数QTL分别位于A03、A07、C01、C04和C06染色体, 单个QTL解释5.95%~8.14%的表型变异。将156个拟南芥株高相关基因、10个拟南芥第1分枝高相关基因和148个拟南芥分枝数相关基因与QTL对应置信区间序列进行同源比较分析(E<1E-20), 分别找出了20个株高候选基因、3个第1分枝高候选基因以及12个分枝数候选基因。2个环境中在A07染色体上重复检测到的QTL置信区间检测到与株高相关的候选基因 ATGID1B/GID1B和 WRI1, A08染色体上重复检测到的QTL置信区间检测到 SLR/IAA14和 AXR2/IAA72个与株高相关的候选基因。在具有部分置信区间重叠的 q2013FBH-C05-1和 q2014FBH-C05-2区间均检测到第1分枝高候选基因 PHT1;8, 在A03和C06染色体上的QTL置信区间内, 分别检测到4个分枝数候选基因, 匹配E值介于0~3E-56之间。
Plant height, the first branch height and branch number are important agronomic traits in rapeseed. In our study, QTL mapping of plant height, the first branch height and branch number in Brassica napus was tested by using the high density SNP genetic map constructed from the high generation RIL population with the rapeseed 60K chip array. The reference SNP genetic map contains 2795 SNP markers, covering 1832.9 cM with an average distance of 0.66 cM in B. napus genome. Totally, 24 putative QTLs were identified for plant height, the first branch height and branch number by using the complex interval mapping. The phenotypic variation was explained by individual QTL ranged from 5.00% to 15.26% for 11 QTLs of plant height, from 5.04% to 12.99% for seven QTLs of the first branch height, and from 5.95% to 8.14% for six QTLs of branch number. We collected 156 genes associated with plant height, 10 genes associated with the first branch height and 148 genes associated with branch number in Arabidopsis thalianaand searched the homology region of the QTL confidence interval E-value<1E-20 to screen the possible candidate genes. We found 20 genes associated with plant height, three genes associated with the first branch height and 12 genes associated with branch number. The plant height candidate genes ATGID1B/GID1B and WRI1 were found in QTL confidence interval on A07 and SLR/IAA14 and AXR2/IAA7 were found in QTL confidence interval on A08 in 2013 and 2014. The first branch height candidate gene PHT1;8 was found in QTLs q2013FBH-C05-1 and q2014FBH-C05-2 that had an overlapping confidence interval. Moreover, we found four genes associated with branch number that E-value from 0 to 3E-56 on A03 and C06, respectively.
油菜(Brassica napusL.)属于十字花科芸薹属, 为世界四大油料作物之一, 菜籽是仅次于大豆的第二大植物油来源[1]。影响油菜产量的农艺性状主要有株高、有效分枝数、分枝部位以及开花时间等。由于油菜农艺性状大多为数量性状, 由微效多基因控制, 表现连续变异, 受环境的影响较大, 不能依照质量性状的处理方法将单个基因的效应区分开, 因此单独依靠传统育种方法和技术在现有基础上研究尚未有大的突破[2]。
DNA分子标记和QTL作图等生物技术的发展为复杂的数量性状的研究提供便利。Li等[3]利用SRAP、SSR、AFLP标记以及功能标记对甘蓝型油菜F2群体在2个环境下同时进行了QTL定位, 检测到12个与一次有效分枝数相关的QTL, 单个QTL能够解释5.47%~27.39%的表型变异。Chen等[4]利用甘蓝型油菜2个群体(DH和IF2)定位了6个分枝数QTL, 其中位于13连锁群上的42.0~55.6 cM的QTL有较大的效应。易斌等[2]利用RFLP、RAPD、SSR以及SRAP等分子标记技术共检测到与油菜产量及其相关性状有关的QTL 17个。其中与株高相关的3个分别位于第4、第9和第10连锁群上, 对性状的解释率为9.42%~17.58%; 与分枝部位有关的4个分别位于第4、第6和第7连锁群上, 其中Bp1和Bp2均位于第4连锁群, 对性状的解释率为8.13%~ 15.20%; 与一次有效分枝有关的2个分别位于第1、第4连锁群上, 对性状的解释率为15.29%~19.58%。张倩[5]利用SSR标记检测到4个与株高相关的QTL, 分别位于LG1、LG2连锁群上, 可解释表型变异的8.54%~17.04%; 与分枝高度相关的5个, 分别位于LG3、LG5、LG8连锁群上, 可解释表型变异的3.92%~11.82%; 与分枝数相关的1个, 位于LG3连锁群上, 可解释表型变异的11.08%。Zhang等[6]利用复合区间作图法, 在第4和第21连锁群上共检测到3个与株高相关的QTL, 每个QTL分别解释28.60%、23.85%和11.11%的表型变异; 在第11、第13、第20和第21连锁群上共检测到4个与分枝高度相关的QTL, 单个QTL能解释8.54%~54.59%的表型变异。
SNP (single nucleotide polymorphism)是基于基因组核苷酸水平上单碱基插入、缺失、转换和颠换引起的多态性差异而开发的DNA标记[7]。随着甘蓝型油菜全基因组测序的完成[8]以及高通量SNP芯片技术的完善和成本的下降, SNP芯片技术将成为油菜遗传研究的热点之一。本研究利用油菜60K芯片构建的高密度SNP遗传连锁图谱, 对株高、第1分枝高和分枝数进行QTL定位分析, 利用甘蓝型油菜基因组数据库序列, 根据QTL区间物理位置及拟南芥基因组数据库信息筛选可能的候选基因。尽可能将数量性状定位结果明确在特定的物理位置甚至候选基因, 有利于不同材料之间定位结果的比较分析及候选基因的克隆分析。
以甘蓝型黄籽油菜GH06作母本, 甘蓝型黑籽型双低油菜P174作父本配制杂交组合, F2通过单粒法再连续自交至F11构建的重组自交系群体, 随机选取其中的172个材料进行SNP标记分析, 构建高密度SNP遗传连锁图谱。所有材料均由重庆市油菜工程技术研究中心提供。
2012年9月至2013年5月(性状统计记录为2013)与2013年9月至2014年5月(性状统计记录为2014)将亲本及重组自交系群体种植于重庆市北碚区歇马镇油菜基地。随机区组设计3个重复, 每个小区2行, 每行10株。行距0.40 m, 株距0.24 m。田间管理同常规生产, 确保所有样本的外部生长环境一致, 待成熟后进行数据统计。
油菜成熟后, 每个株系随机选取5株测量株高、第1分枝高(有效分枝)、分枝数, 取平均值。株高为从地面到植株最高部位的距离(cm); 第1分枝高为地面到第1有效分枝的高度(cm); 分枝数为该植株所有有效的一次分枝数。
本高密度SNP遗传图谱包含2795个SNP多态性标记位点, 图谱总长1832.9 cM, 相邻标记间平均距离为0.66 cM [9]。采用QTL分析软件Windows QTL Cartographer 2.5[10]及复合区间作图(composite interval mapping, CIM)法对该群体的株高、第1分枝高、分枝数进行QTL定位及效应检测[11]。CIM分析时, 选取1 cM的步长(walking speed), 按照假定检测10和Zmapqtl模型6, 选取参数为1000次回归, 显著水平为0.01。LOD≥ 3.0时, 即认为该区间可能存在一个QTL。运行软件后可同时给出各QTL的加性效应和解释的表型变异。按照McCouch等[12]的方法对检测到的QTL命名, 以“ q” 加上性状再加染色体编号表示, 字体为斜体。同一性状在染色体相同的位置检测重复的QTL, 且加性效应方向一致, 认为是同一QTL。
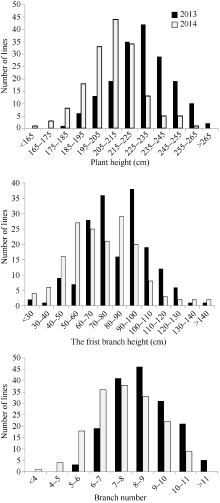
如表1和图1所示, RIL群体中株高、第1分枝高和分枝数在2年的均值差异都较小, 表明其是多位点控制的典型数量性状, 且位点间加性效应在同一亲本中具有不同的方向。各性状在RIL群体中均呈连续分布和双向超亲分离, 表明这些性状均为多基因控制的数量性状, 其偏斜度和峰值均小1, 适合于QTL分析。
相关分析表明(表2), 同一性状在2013年和2014年表现均呈极显著正相关, 且相关系数较大, 说明株高、第1分枝高和分枝数性状遗传稳定。性状间的相关分析还表明, 在2个生长周期里, 第1分枝高和株高呈极显著正相关, 相关系数分别为0.468和0.451; 分枝数与株高呈显著负相关或未达到显著水平。同时第1分枝高和分枝数在2年里均呈极显著负相关, 相关系数分别为-0.590和-0.475。
利用软件Windows QTL Cartographer 2.5对2年的株高、第1分枝高、分枝数进行QTL分析, 共检测到24个QTL (表3和图2)。其中11个株高QTL, 阈值3.02~7.51, 单个QTL解释的表型变异为5.00%~15.26%。在2年内检测到对株高有显著效应的共同QTL位点(q2013PH-A07和q2014PH-A07-1、q2013PH-A08和q2014PH-A08), 分别位于A07和A08染色体上, 加性效应值均为负, 说明增效基因来自父本P174, 在C06染色体上两年内也检测到一个具有重叠置信区间的QTL (q2013PH-C06和q2014PH- C06), 加性效应值大于0, 说明增效基因来自母本GH06。对于第1分枝高在2年内检测到7个QTL, 阈值为3.54~5.89, 单个QTL解释的表型变异为5.04%~ 12.99%, 分别位于C05、C06和A06染色体上。对于分枝数在2年内共检测到6个QTL, 阈值为3.20~4.54, 单个QTL解释的表型变异为5.95%~8.14%。
| 表1 亲本及重组自交系群体3个农艺性状在2年的表型分析 Table 1 Analysis of three agronomic traits in the two parents and RIL population in 2013 and 2014 |
| 表2 甘蓝型油菜重组自交系群体农艺性状在2013年和2014年的相关性分析 Table 2 Correlation analysis for agronomic traits from B. napus RIL population in 2013 and 2014 |
本研究将24个QTL置信区间序列与156个株高基因、10个第1分枝高基因和148个分枝数基因分别进行比对, 其中有11个与株高相关的QTL共检测到20个候选基因, 匹配E值介于0~9E-24之间, 2个与第1分枝高相关的QTL共检测到3个候选基因, 5个与分株数相关的QTL共检测到12个候选基因, 匹配E值介于0~3E-56之间。其中在q2013PH-A06和q2013PH-A10置信区间内分别筛出3个株高候选基因, q2013PH-A07、q2013PH-A08、q2014PH-A01、q2014PH-A07-1、q2014PH-A08以及q2014PH-C06置信区间内分别筛出2个株高基因。株高基因IAA14/ SLR和IAA7/AXR2在2年重复检测到的QTL (q2013PH-A08和q2014PH-A08)区间内均检测到, 在2年内具有重叠置信区间的QTLq2014PH- A07-1和q2013PH-A07区间内均检测到株高基因ATGID1B/ GID1B和WRI1。第1分枝高候选基因PHT1; 8在具有部分置信区间重叠的QTL q2013FBH-C05-1和q2014FBH-C05-2中均检测到, q2013BN-A03和q2014BN-C06区间内分别检测到4个分枝数候选基因。比对结果列于表4, 候选基因的功能列于表5。
油菜的农艺性状主要包括株高、有效分枝数、第1分枝高度、千粒重、开花时间、主花序长、有效角果总数、果长、每果粒数等。株高作为一个重要的性状受到了育种家的重视, 这不仅因为株高在油菜产量形成过程中具有重要的间接支配作用, 而且株高直接影响能否有效机械收割。分枝不仅是油菜角果的重要载体, 还影响其形态建成, 对油菜产量具有十分重要的作用。一次有效分枝数作为角果数的重要载体, 对油菜产量起着重要的作用, 在一定范围内增加油菜分枝数可以显著增加全株角果数, 进而提高单株产量[44, 45]。对于油菜农艺性状与产量的关系分析, 已有大量报道, 虽然结果不尽一致, 但普遍认同单株角果数与分枝数对产量的贡献最大[46, 47, 48, 49]。
对油菜株高、第1分枝高以及分枝数的遗传分析大多停留在经典数量遗传学研究方面, 对油菜分枝数等农艺性状的遗传机制了解较少[50]。相对于其他作物, 甘蓝型油菜农艺性状QTL定位研究开展比较晚, 而且到目前为止, 只报道了为数不多的农艺性状QTL定位研究和少量的QTL[51, 52, 53, 54, 55], 而且定位结果不尽一致, 这也从侧面表明油菜农艺性状的复杂性。因此, 研究株高、第1分枝高及分枝数具有重要的现实意义。利用高密度SNP遗传图谱已经对甘蓝型油菜的种皮纤维素[9]、种子硫苷含量[56]及盐、旱胁迫下种子发芽率[57]进行了准确定位。本研究利用高密度SNP遗传图谱对甘蓝型油菜株高、第1分枝高及分枝数进行QTL分析, 均在同一标记区间或紧邻的染色体区域内重复检测到一个或多个QTL, 但其效应值均较小, 没有主效基因。
| 表3 利用复合区间作图法检测到的株高(PH)、第1分枝高(FBH)和分枝数(BN)在2个环境中的QTL Table 3 Putative QTL detected by composite interval mapping for plant height (PH), the first branch height (FBH), and branch number (BN) in two environments |
Wei等[58]利用258个DH株系以SSR和SRAP标记技术共检测到20个与株高相关的QTL, 分布在N3、N10、N13、N16和N17连锁群上, 单个QTL解释6.2%~ 12.0%的表型变异; 检测到10个与第1分枝高相关的QTL, 分布于N2、N8、N16和N17连锁群上, 单个QTL解释4.3%~16.2%的表型变异。本研究除在普遍检测到株高QTL的染色体A07、A08和C06上[13, 14, 15, 59, 60, 61]检测到株高相关的QTL外, 还在A01和A10染色体上各检测到1个株高QTL; 在A06、C05及C06染色体上检测到7个与第1分枝高相关的QTL; 在A03和C06染色体上检测到的分枝数QTL与前人研究结果[3, 4, 13, 14, 15, 59, 60, 61]一致, 另外在A07、C01及C04染色体上检测到3个与分枝数相关的QTL, 单个QTL解释的表型变异为6.13%~ 8.14%。
Tuberosa等[62]认为性状相关表现在QTL分析结果上, 可能存在控制不同性状QTL之间的紧密连锁或同一QTL位点控制不同的性状现象即一因多效。本研究中, 在C06染色体上检测到6个与株高、第1分枝高及分枝数相关的QTL, 其中q2013PH-C06和q2013FBH-C06-1的区域紧密连锁, 佐证了株高与第1分枝高呈极显著正相关。而位于C06染色体上控制株高和分枝数的QTL增效基因相反, 印证了株高与分枝数呈显著负相关。前人对油菜的株高、第1分枝高和分枝数等农艺性状的QTL定位结果不尽一致, 这是由于这些性状由微效多基因控制, 受环境影响较大, 因此筛选出控制这些农艺性状的候选基因显得极有意义。Shi等[15]在低磷和最佳施磷量条件下, 用30个GBM标记在检测到的QTL的置信区间发现19个候选基因。其中在缺磷和最适条件下均在A09染色体上的QTL置信区间检测到1个控制株高的候选基因(LPR1), 分别在A01和A06染色体上的QTL置信区域检测到与分枝数有关的候选基因PAP17、FBHLH32和PHT1; 8, 在A01、A02、A03、A04、A09和C07染色体上的QTL区域共检测到10个与第1分枝高相关的候选基因。Ding等[14]在70个QTL置信区域内检测到136个与甘蓝型油菜农艺性状相关的候选基因, 其中46个与株高相关, 30个为分枝数的候选基因, 此结果与Shi等[15]搜索的部分结果一致。本研究利用Shi等[13]、Ding等[14]和Shi等[15]搜索或检测到的相关候选基因与本文检测到的QTL置信区间序列进行同源比对, 检测到20个株高候选基因、3个第1分枝高候选基因和12个分枝数候选基因。2个环境中在A07染色体上重复检测到的QTL置信区间检测到与株高相关的候选基因ATGID1B/GID1B和WRI1, A08染色体上重复检测到的QTL置信区间检测到SLR/IAA14和AXR2/IAA72个与株高相关的候选基因。在具有部分置信区间重叠的QTL q2013FBH- C05-1和q2014FBH-C05-2区间均检测到第1分枝高候选基因PHT1; 8, 与Shi等[15]检测到该基因的位置(A01和A06染色体上)存在差异。在位于前人检测到分枝数QTL的A03和C06染色体上[3, 4, 13, 14, 15, 59, 60, 61]的QTL置信区间内, 我们分别检测到4个分枝数候选基因, 匹配E值介于0~3E-56之间, 暗示这两个QTL极有可能为控制分枝数的遗传位点, 其结果有待进一步验证。检测到株高、第1分枝高及分枝数的候选基因较少, 可能与QTL的贡献率小有关, 检测到第1分枝高相关候选基因较少的原因还可能是在本研究中用于搜索的第1分枝高相关基因较少。
 | 图2 甘蓝型油菜株高、第1分枝高及分枝数QTL在SNP连锁群上分布图Fig. 2 Putative QTL locations of plant height, the first branch height, and branch number on the SNP genetic map |
| 表4 甘蓝型油菜基因组中QTL置信区间候选基因与拟南芥相关基因的比对 Table 4 Alignment of candidate genes in QTL confidence interval in B. napuswith the related genes in Arabidopsis thaliana |
| 表5 拟南芥中株高(PH)、第1分枝高(FBH)和分枝数(BN)候选基因功能 Table 5 Functions of candidate genes related to plant height (PH), the first branch height (FBH), and branch number (BN) in A. thaliana |
同一环境下株高与第1分枝高呈极显著相关, 与分枝数呈负相关, 第1分枝高与分枝数在2个环境中均呈极显著负相关。在2个环境下重复检测到3个与株高相关的QTL, 分别位于A07、A08和C06染色体上, 在A07染色体上重复检测到的QTL置信区间检测到2个与株高相关的候选基因ATGID1B/GID1B和WRI1, 在A08染色体上的QTL置信区间检测到3个与株高相关的候选基因(SLR/ IAA14、AXR2/IAA7和BIN2/DWF12); 第1分枝高、分枝数各重复检测到1个QTL, 分别位于C05和C06染色体上。位于A03和C06染色体上的QTL q2013BN-A03和q2014BN-C06分别检测到4个分枝数候选基因, 极有可能为控制分枝数的遗传位点。
The authors have declared that no competing interests exist.
作者已声明无竞争性利益关系。The authors have declared that no competing interests exist.
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
|
| [45] |
|
| [46] |
|
| [47] |
|
| [48] |
|
| [49] |
|
| [50] |
|
| [51] |
|
| [52] |
|
| [53] |
|
| [54] |
|
| [55] |
|
| [56] |
|
| [57] |
|
| [58] |
|
| [59] |
|
| [60] |
|
| [61] |
|
| [62] |
|