第一作者联系方式: E-mail:zhujifeng0224@163.com
菜豆普通细菌性疫病是世界上危害普通菜豆生产的最严重的病害之一。龙芸豆5号是我国黑龙江省的主栽品种, 对菜豆普通细菌性疫病表现出良好的抗性。为定位来源于龙芸豆5号的抗性基因, 本研究构建了包含785个单株的F2分离群体。基于该群体构建了一张包含206个SSR标记, 总长度1648.42 cM, 标记间平均遗传距离8.00 cM的遗传图谱。图谱包含12个连锁群, 各连锁群平均长度137.37 cM, 连锁群上标记数量3~35个。结合温室表型鉴定结果, 采用QTL IciMapping v4.0软件的完备区间作图法进行QTL定位和效应估计。接种14 d后在Pv06染色体上检测到一个抗病QTL。该位点位于标记 p6s249与 p6s183之间, 加性效应值为0.44, 说明增效基因来源于龙芸豆5号, LOD值为5.93, 表型贡献率为4.61%, 该抗病QTL的效应值相对较低, 将在培育稳定持久的抗菜豆普通细菌性疫病的品种中发挥作用。最后, 对抗性基因紧密连锁的11对SSR引物与菜豆普通细菌性疫病抗性的关联分析表明, SSR标记 p6s249与菜豆普通细菌性疫病抗性极显著关联( P<0.001), 该标记可用于抗病分子育种。
Common bacterial blight disease is a serious disease affecting the production of common bean ( Phaseolus vulgaris L.) worldwide. The common bean germplasm Longyundou 5 is the main cultivar carrying common bacterial blight resistance gene in Heilongjiang province. To study the genetic mechanisms behind this resistance, we constructed 785 F2 plants from Longyundou 5. Linkage analysis was performed on this population by using SSR markers. A linkage map covering 1648.42 cM with an average marker distance of 8.00 cM among 206 SSR markers was constructed. This map contained 12 linkage groups with a mean group length of 137.37 cM and the number of loci ranging from three to thirty-five. Combined with the results of phenotypic evaluation, QTL analysis was performed by the inclusive composite interval mapping method with QTL IciMapping v4.0. A QTL was detected between p6s249 and p6s183 on chromosome Pv06, with an additive effect of 0.44, which means the favourable gene of this locus is from Longyundou 5. The LOD score of the QTL was 5.93 and the total phenotypic variation at 14 days after inoculation was 4.61%. These results showed the effect of the QTL was low, which will play an important role in breeding durable resistant varieties of common bean. Association analysis between 11 SSR primers linked with common bacterial blight resistance and the disease rating showed that SSR marker p6s249 is excellently associated ( P<0.001) with common bacterial blight resistance and can be used for marker assisted selection.
普通菜豆(Phaseolus vulgaris L.)是人类生活中重要的食用豆类之一, 富含蛋白质、碳水化合物、维生素等人体所必需的营养成分[1]。2014年全世界普通菜豆籽粒产量为2500万吨, 占食用豆总产量的50%左右(FAOSTAT 2015)。然而, 由地毯草黄单胞杆菌菜豆致病变种(Xanthomonas axonopodis pv. phaseoli, Xap)[2]所引起的菜豆普通细菌性疫病(common bacterial blight, CBB)严重威胁着普通菜豆的生产, 已成为全世界普通菜豆生产中的主要病害[3]。菜豆普通细菌性疫病为种传细菌性病害, 在普通菜豆全生育期内皆可发生, 危害寄主植物的茎、叶、嫩荚和种子, 致使产量及品质下降; 由该病导致的普通菜豆产量损失一般为20%~60%, 严重时高达80%, 甚至绝产[4]。这就迫切要求我们鉴定抗菜豆普通细菌性疫病的优异种质, 并从中挖掘优异基因或有效功能标记, 将其有效应用于普通菜豆抗病分子育种。
迄今为止, 国内外已报道定位了24个抗菜豆普通细菌性疫病的QTL [3, 5, 6, 7, 8, 9], 分布于普通菜豆的11条连锁群上[3, 6, 9]。其中, 位于染色体Pv07上的菜豆普通细菌性疫病抗病位点最多, 有5个[10, 11, 12]; 而在染色体Pv01 [11]、Pv04 [7]、Pv06 [7]和Pv10 [10, 11]上各检测到的一个抗病位点。在已定位的抗病位点中, 对SAP6[13]、BC420[7]和SU91[14]的研究较为深入。SAP6来源于普通菜豆(Montana 5)[13], 而BC420、SU91均来源于普通菜豆的近缘种宽叶菜豆(PI 319443)[15]。SAP6位于染色体Pv10[13], 可解释35%的表型变异[13]。BC420位于染色体Pv06 [7], 可解释62%~63%的表型变异[12]。SU91位于染色体Pv08[14], 可解释14%~ 17%的表型变异[16]。由于已报道的24个抗性基因的分子标记多为显性标记如RAPD、SCAR标记等, 不能鉴别杂合子和纯合子, 限制了其在抗病分子育种中的应用。在已定位的抗病基因中, 仅针对BC420和SU91基因开发了共显性标记且被广泛应用于普通菜豆的抗病分子育种[17]。为此, 需要定位克隆新的菜豆普通细菌性疫病抗性基因, 并开发共显性标记, 为抗病分子辅助选择育种提供更多、更有效的分子标记。
普通菜豆龙芸豆5号在温室和田间对我国菜豆普通细菌性疫病病菌Xap菌株XS2均表现出较好的抗性。目前, 关于龙芸豆5号的抗病性遗传分析研究尚未报道。本研究利用龙芸豆5号与感病材料龙芸豆4号配置的F2杂交群体, 以SSR分子标记定位来源于龙芸豆5号的抗病基因, 并为抗病分子育种提供有效的分子标记。
普通菜豆抗病品种龙芸豆5号(龙5)和感病品种龙芸豆4号(龙4)为黑龙江省农业科学院的2个育成品种, 由中国农业科学院作物科学研究所提供。以龙5为父本、龙4为母本, 通过杂交和自交获得F2分离群体共785个单株, 用于表型鉴定、遗传分析和抗病QTL定位。
取保存于-80℃超低温冰箱的Xap菌株XS2于牛奶吐温培养基[18]上活化4~5 d (28℃± 2℃), 然后转入牛肉膏蛋白胨液体培养基中振荡培养48 h (200转 min-1, 28℃± 2℃), 用灭菌蒸馏水稀释至菌液浓度为1× 108 cfu mL-1, 用于接种。
以亲本材料龙5为抗病对照、龙4为感病对照, 将亲本与F2群体播种于口径为23 cm × 18 cm × 18 cm的花盆中, 每盆5粒, 每亲本材料4盆, 控制温室白天温度为28℃± 2℃、晚上温度为20℃± 2℃, 待第一片三出复叶完全展开后采用针刺叶片法接种[19]。接种14 d后调查接种植株叶片的发病情况, 参照Zapata等[20]评级标准, 调查记录每株接种叶片的发病严重度, 并计算亲本龙5和龙4的平均发病级别, 利用软件SAS 9.1对数据进行统计分析。
采用改良CTAB法[21]提取龙5、龙4及F2单株的基因组DNA。用于遗传分析的SSR引物包括两部分, 共计3186对。第1部分来源于已发表文献[22, 23], 计381对; 第2部分是本实验室基于普通菜豆全基因组序列[24]所开发, 计2805对。上述SSR引物全部用于亲本间的多态性分析, 选取在亲本间有明显多态性且带型清楚的引物分析F2群体的遗传连锁性。
PCR扩增体系为15 µ L, 含20 ng模板DNA、0.2 µ mol L-1正反引物(Invitrogen, USA)、0.25 mmol L-1 dNTPs (dATP、dCTP、dGTP与dTTP)、1.5 µ L的10× Taq缓冲液(含1.5 mmol L-1 Mg2+)以及1 U DNA聚合酶。PCR扩增程序为95℃预变性5 min; 95℃变性30 s, 53℃退火45 s, 72℃延伸45 s, 循环35次; 最后72℃延伸10 min。PCR扩增产物经8%非变性聚丙烯酰胺凝胶电泳检测, 银染法[25]染色后观察记录结果。
利用QTL IciMapping v4.0[26]构建F2群体的遗传连锁图谱, 设置LOD≥ 3.0、最大遗传距离30 cM, 选用Kosambi作图函数[27]作图。参照普通菜豆全基因组序列[24], 将连锁群与染色体一一对应。
结合构建的遗传连锁图谱和表型鉴定结果, 采用完备区间作图法(inclusive composite interval mapping, ICIM)[28]检测龙5中LOD值≥ 3.0的抗性基因位点。
利用TASSEL 2.1软件[29]中一般线性模型(general linear model, GLM)对目标区段所在染色体上的SSR标记与CBB抗感结果进行标记-性状关联分析, 结合QTL检测结果确定与抗病性紧密关联的位点。
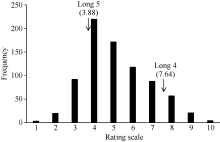
接种14 d后, 抗病亲本龙5接菌叶片没有出现感病症状, 表现出较强抗性(图1), 平均发病级别为3.88 (图2); 感病亲本龙4接菌叶片的接种区和对照区均枯萎变色(图1), 平均发病级别为7.64 (图2), 表现为感病。F2群体表现出明显的抗感分离, 最大值超过了高值亲本, 最小值低于低值亲本, 表明性状同时具有正向和负向超亲优势, 且呈连续分布(图2), 其偏度(Skewness)值为0.46, 峰度(Kurtosis)值为-0.30, 二者的绝对值均小于0.5, 符合正态分布, 适合进行QTL定位分析。
利用3186对SSR引物扩增亲本龙5和龙4基因组DNA表明, 有228对引物在亲本间具有多态性。利用这些多态性引物对785个F2单株进行遗传连锁分析, 获得含206个SSR标记的遗传连锁图谱, 共分12个连锁群, 其中最短连锁群为Pv02, 仅含3个标记、最长连锁群为Pv08, 含35个标记(图3)。依据标记的染色体信息, 确定每个连锁群对应的染色体, 染色体Pv03由于标记密度不够而断开为2个连锁群(Pv03a与Pv03b), 其余连锁群与其染色体一一对应(图3)。该遗传图谱总长度为1648.42 cM, 平均遗传距离为8.00 cM; 标记间最短遗传距离为0.60 cM, 位于Pv06上的p6s267与p6s123标记之间; 标记间最大遗传距离为36.92 cM, 位于Pv06上的p6s126与p6s277标记之间(图3); 标记密度最大的是Pv04连锁群, 其标记间平均遗传距离只有5.65 cM。
结合遗传连锁图谱和F2表型鉴定结果, 利用软件QTL IciMapping v4.0进行ICIM定位分析, 接种后14 d仅在Pv06染色体上检测到一个抗病QTL, 该位点位于分子标记p6s249与p6s183之间(图4-A), 其LOD值为5.93 (图4-B), 共解释4.61%的表型变异。此外, 在Pv06染色体上检测到的抗病QTL加性效应值为0.44, 说明增效基因来源于龙芸豆5号, 且该抗病QTL能增强龙芸豆5号对菜豆普通细菌性疫病的抗性。
利用GLM法对抗病QTL所在连锁群(Pv06)上的所有标记关联性分析表明, 该连锁群上有11对SSR引物与CBB发病等级显著相关(P< 0.05), 其中个SSR标记p6s302、p6s281、p6s267、p6s249、p6s210、PVBR163与CBB抗性的相关性达到P< 0.001的显著水平(表1)。结合本研究所检测到的抗性QTL的侧翼标记(p6s249与p6s183)(图4-A), 发现与菜豆普通细菌性疫病抗性位点紧密关联的标记为p6s249, 该引物在G19833基因组中预扩增目标片段为216 bp (表1)。p6s249标记在龙5基因组中扩增出的目标片段位于201~238 bp之间, 在龙4扩增出的片段位于238~242 bp之间(图5)。
由地毯草黄单胞杆菌菜豆致病变种[2]引起的菜豆普通细菌性疫病是影响全世界菜豆生产的一种主要病害[3]。研究表明菜豆普通细菌性疫病的抗性是受少数主效基因控制的数量性状[8, 12]。目前, 已报道的24个与CBB抗性相关的QTL中, 位于染色体Pv07上有5个[10, 11, 12], Pv02上4个[7, 10, 11], Pv08上3个[5, 14], Pv03[7, 10]、Pv05[7]、Pv09[11]和Pv11[10]上各2个, Pv01[11]、Pv04[7]、Pv06[7]和Pv10[10, 11]上各1个。这些抗病QTL中, 位于Pv06染色体上的(BC420)因其具有较高贡献率而得到深入研究[6, 17]。
本研究在Pv06染色体上检测到一个抗Xap菌株XS2的QTL, 该抗病位点位于分子标记p6s249与p6s183之间(图4-A), LOD值为5.93 (图4-B), 贡献率为4.61%, 抗病效应值较低, 可能将在培育稳定持久的抗菜豆普通细菌性疫病的品种中发挥作用。
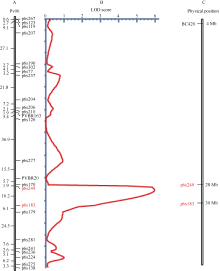 | 图4 F2群体中抗病QTL的定位 B: 红线代表在Pv06上检测到的抗病QTL的LOD值。Fig. 4 Mapping of QTL for CBB resistance in F2 population B: The red line means the LOD score of the major QTL for CBB resistance on chromosome Pv06. |
| 表1 SSR引物与CBB抗性间的关联分析 Table 1 Testing of association between SSR primers and CBB resistance |
依据SSR引物p6s249与p6s183在G19833物理图谱上的位置可初步断定本研究获得的抗病QTL位于Pv06染色体28~30 Mb的物理距离处(图4-C)。根据Shi等[17]所确定的位于Pv06染色体上的抗性基因BC420的候选基因序列比对分析, 该抗病位点位于第6染色体4 Mb的物理距离处, 与本研究检测到的抗病位点相距较远(图4-C), 因此我们所发现的抗病位点可能是一个来源于普通菜豆的新抗病位点。
利用与目的基因紧密连锁的DNA标记进行分子标记辅助选择有利于加快作物育种进程、提高育种效率[30]。目前, DNA标记已被广泛应用于水稻[31]、玉米[32]、小麦[33]等作物的分子育种。在众多的DNA标记中, SSR标记因其具有多态性高、共显性、数量丰富、信息量高、重复性好、DNA需求量低、易检测、简单经济等优点, 成为作物分子标记辅助选择育种中最好用的一种标记类型[34]。此外, 由DNA的特殊序列标记(如RFLP、RAPD等)转换而来的STS、SCAR和SNP等标记也同样被广泛应用于分子标记辅助选择育种[34]。分子标记辅助选择育种技术在菜豆普通细菌性疫病抗病育种中也得到应用, 特别是3个重要的SCAR标记BC420、SU91和SAP6已被用于菜豆普通细菌性疫病的MAS育种[6, 35]。然而, 由于SCAR标记同其他共显性标记一样不能区分杂合位点, 在一定程度上限制了它的应用。Shi等[17]开发了共显性候选基因标记BC420-CG14和SU91- CG11来代替SU91和BC420用于CBB抗病育种。本研究获得了一个与CBB极显著关联的SSR标记p6s249, 该标记所在位点远离BC420, 为一个新抗病位点连锁的标记; 此外, 该标记来源于普通菜豆, 更易于普通菜豆育种选择, 所以p6s249可能有助于今后的MAS抗病育种。
定位了1个普通菜豆抗菜豆普通细菌性疫病QTL, 该位点位于标记p6s249与p6s183之间, 共解释4.61%的表型变异率, 效应值较小, 将在培育稳定持久抗菜豆普通细菌性疫病品种中发挥作用。p6s249与菜豆普通细菌性疫病抗性极显著关联(P< 0.001), 可被用于抗病分子育种。
The authors have declared that no competing interests exist.
作者已声明无竞争性利益关系。
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|