内源小干扰RNAs (small interfering RNAs, siRNAs)和DNA甲基化在植物生长发育和适应环境胁迫中调控基因的表达。对于植物来说, 镉(Cadmium, Cd)是一种非必需且具有毒性的元素。为研究DNA甲基化和siRNAs在水稻(
In plants, as in other eukaryotes, endogenous small interfering RNAs (siRNAs), a class of small non-coding RNAs, and DNA methylation regulate gene expression in developmental processes and adaptating to environmental stresses, including Cd stress. Cadmium (Cd) is a non-essential heavy metal and highly toxic to plants. To investigate the regulatory role of siRNAs and DNA methylation on genes involved in heavy metals transport, we compared these genes’ expression profiles between a high Cd-accumulating rice (
镉(cadmium, Cd)是一种毒性极强的重金属[ 1]。近年来, 熔炼、采矿和含Cd化肥的施用等造成大量农田Cd污染[ 2]。水稻根系易从土壤中吸收Cd, 并向地上部分转移后积累在稻米中, 造成稻米Cd含量超标[ 1, 3]。Cd可通过食物链进入人体, 对人类健康造成严重威胁[ 4]。
研究认为, Cd转运子和Cd2+螯合蛋白参与水稻体内Cd的积累[ 5, 6, 7, 8, 9]。 OsLCD基因在水稻根部维管束和叶片韧皮部伴胞中表达, 参与稻米中Cd的积累[ 7]。 OsLCT1基因在水稻叶片和节的维管组织中表达[ 1]。OsLCT1蛋白是定位于细胞膜的蛋白, 参与Cd从细胞内部向外界的排放[ 1]。植物体内编码Cd2+螯合蛋白的基因, 如金属硫蛋白基因 OsMT1、富含半胱氨酸多肽基因 OsDCT1和植物螯合肽基因 OsPCs, 参与其体内Cd2+的螯合和转移[ 5, 10]。
近年来的研究证实植物Cd抗性蛋白(plant cadmium resistance protein, PCR)和植物Cd抗性相关, 它含有CCXXXCPC保守氨基酸序列, 属于膜蛋白家族[ 11]。该蛋白家族成员通常形成同源聚合体, 参与Cd2+从细胞内部向外界的转移[ 8, 9, 12], 尤其在植物根部对Cd2+的排放和Cd从植物地下部向地上部的转移过程中发挥重要作用[ 9, 12]。
虽报道有大量的Cd积累相关基因, 但其表达的分子调控机制尚不明确。和其他真核生物一样, 在应对环境胁迫过程中, 包括重金属胁迫, 在植物中基因的表达受到内源小干扰RNA的调控[ 13, 14, 15, 16, 17]。siRNAs是通过RNA酶III家族成员DICER剪切双链RNA形成的一类长度为20~25个核甘酸(nt)具有基因沉默作用的RNA双链单位[ 18]。siRNAs的一条链选择性结合到Argonaute (AGO)蛋白从而形成RNA介导的沉默复合物(RNA-induced silencing complex, RISC)[ 19], AGO剪切与siRNA互补配对的靶mRNA[ 20], 最终导致靶基因沉默而失活[ 18]。
植物siRNAs与DNA甲基化相关[ 21]。胞嘧啶甲基化修饰是一种基因沉默机制, 真核生物常用它失活自私DNA重复序列(包括转座子)来保护基因组[ 22]。现已发现植物在发育和适应环境胁迫的过程中也使用DNA甲基化来调控内源基因的表达[ 21, 22, 23, 24, 25, 26, 27]。在很多生物的发育过程中, 转录的基因在基因本体区域(外显子区域或者内含子区域)和启动子区域都出现高水平的DNA甲基化[ 28, 29, 30, 31, 32], 而且DNA甲基化是高度动态的[ 33]。DNA甲基化水平的动态变化参与基因的表达调控。
本研究采用qRT-PCR技术比较了不同生长发育期Cd高积累水稻品种(秀水110)和Cd低积累水稻品种(秀水11)叶片中Cd积累相关基因的表达情况来阐述水稻体内Cd积累的分子调控机制。研究表明2个水稻品种叶片中除 OsPCR1外, 其他Cd积累相关基因的表达不存在显著的差异。结果分析表明, 和 OsPCR1基因第2个外显子区域匹配的siRNA和该外显子区域的DNA甲基可能共同调控 OsPCR1基因的表达。
水稻品种秀水11 ( Oryza sativa L. subsp. japonica cv. Xiushui 11)和秀水110 ( Oryza sativa L. subsp. japonica cv. Xiushui 110)。在含有3.62 mg kg-1 Cd污染条件下种植的秀水110中, 稻米中Cd的浓度比秀水11高10.38倍[ 34]。这里定义稻米中Cd浓度高的水稻品种为Cd高积累水稻品种, 稻米中Cd浓度低的水稻品种为Cd低积累水稻品种。秸秆中Cd的浓度和稻米中Cd的浓度具有极显著的相关性[ 34]。两品种的Cd积累模式不同, 在苗期秀水11叶片中Cd含量高于秀水110[ 35], 而在成熟期秀水11叶片中Cd含量低于秀水110[ 34]。
选取成熟饱满的水稻种子, 用5%的次氯酸钠溶液消毒15 min, 无菌水漂洗数次, 37℃黑暗培养箱中催芽2 d至种子露白。将露白的种子播于泡沫网纱浮板上, 于在13 h/11 h (白天/黑夜)光周期、28℃/22℃(白天/黑夜)温周期和70%相对湿度可控培养箱中培养。用1/2培养液培养2周, 将水稻幼苗移至含3 L全培养液的直径为15 cm的塑料桶中。每桶种16穴, 每穴1株, 用塑料板分隔各穴。用海绵固定水稻苗, 使其垂直生长。每周换1次培养液, 调节pH值至5.4~5.5之间。
在培养箱中培养30 d之后, 用10 μmol L-1 CdCl2处理60 d测定水稻体内Cd积累情况。在培养箱中培养30 d后, 用5 μmol L-1 CdCl2处理直到营养生长期(Cd处理后1 d)、抽穗前期(Cd处理后35 d)、抽穗期(Cd处理后60 d)、成熟期(Cd处理后105 d)和完全成熟期(Cd处理后130 d)分析水稻叶片中基因表达、siRNA和DNA甲基化。设置对照组, 除不加CdCl2外, 其他实验条件与Cd处理组完全一致。
取叶片在105℃杀青2 h, 在75℃烘72 h。用 6 mL硝酸和2 mL氢氟酸消解400 mg叶片干样品。用石墨炉原子吸收光谱仪(GF-AAS: SperctrAA7000 Shimadzu Inc., Japan)测定样品中Cd的含量。
用TRIzol (Invitrogen, USA)试剂提取营养生长期、抽穗前期、抽穗期、成熟期和完全成熟期水稻倒二叶和倒三叶总RNA。用DNase I (TaKaRa, Japan)去除DNA。参照Ding等[ 36]的方法合成cDNA, 使用oligo(dT)引物和PremixScript (TaKaRa, Japan)逆转录试剂盒进行逆转录, 然后用SYBR Premix Ex Taq (TaKaRa, Japan)在Rotor gene Q (Qiagen, Shanghai, China) qRT-PCR仪检测系统检测, 最后以 OsUBQ5为对照进行标准化定量。PCR条件为95℃ 1 min变性, 然后95℃ 10 s、58℃ 15 s、72℃ 30 s进行45个循环。用Pfaffl法[ 37]计算相对表达丰度。所有试验重复3次以上。引物序列见表1。
| 表1 qRT-PCR所用的引物序列 Table 1 Primer sequences used in qRT-PCR |
RNA提取和DNA去除方法同1.4。设计针对siRNA具有茎环结构的RT引物(5°-GTCGTATCCAG TGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCCTGCA-3°)[ 36]。该引物末端的6个碱基和siRNA的配对, 其余碱基形成颈环结构。颈环引物和siRNA杂交, 用PremixScript (TaKaRa, Japan)逆转录酶进行逆转录。专一的siRNA前引物和后引物加入到PCR反应管, 然后用SYBR Premix Ex Taq (TaKaRa, Japan)在Rotor gene Q (Qiagen, Shanghai, China) qRT-PCR仪检测系统检测, 最后以U6核小RNA为对照进行标准化定量。PCR条件为 95℃ 1 min变性, 然后95℃ 10 s、58℃ 15 s、72℃ 15 s进行45个循环。用于U6核小RNA逆转录的颈环引物为
5°-ATTTGGACCATTTCTCGATTTGT-3°。用于扩增siRNA的前引物是 5'-CAGTGCGTGTCGTGGAGT-3', 后引物是5'-AAAGCGGTCCCAGTACGGCCTGCA-3'; 用于扩增U6 snRNA的前引物是5'-CGATAAAAT TGGAACGATACAGA-3', 后引物是5'-ATTTGGAC CATTTCTCGATTTGT-3'。用Pfaffl法[ 37]计算相对表达丰度。所有试验重复3次以上。
采用CTAB法提取1.4中所描述叶片的基因组DNA[ 38]。用RNase A去除RNA。参照Teixeira等的方法[ 39]检测DNA甲基化水平。使用McrBC酶(New England Biolab)过夜(16 h)酶切基因组DNA (1 μg), 该酶切割发生甲基化修饰的DNA形成DNA碎片, 然后在65℃灭活20 min。qRT-PCR使用相同量(5 ng)
经酶切和未经酶切的基因组DNA, 然后用SYBR Premix Ex Taq (TaKaRa, Japan)在Rotor gene Q (Qiagen, Shanghai, China) qRT-PCR仪检测系统检测, 最后以未经酶切的基因组DNA为对照进行标准化定量。PCR条件为 95℃ 1 min变性, 然后95℃ 10 s、58℃ 15 s、72℃ 30 s进行45个循环。用于扩增 OsPCR1基因外显子2区域的专一性前引物是5'-GCTGCAACTGCGTCTACTCCTGCTT-3', 后引物是5'-CGTGCCATCCGAGGTTCATGTCGAA-3'。结果以相对未甲基化DNA水平表示, 用Pfaffl法计算[ 37]。所有试验重复3次以上。
数据以平均值±标准误表示, 3~8个独立重复试验, 采用配对 t检验检测显著性水平。 P<0.05表示差异显著, P<0.01表示差异非常显著, P<0.001表示差异极其显著。
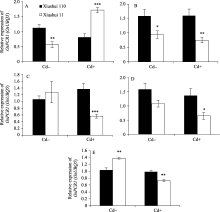
秀水110叶片中Cd的含量为30.9±0.8 mg kg-1,而秀水11叶片中Cd的含量为23.5±0.3 mg kg-1 ( P< 0.001, t检验)。为研究水稻体内Cd积累的分子调控机制, 采用qRT-PCR方法对编码Cd转运子和Cd2+螯合蛋白的基因进行了表达检测。根据水稻发育的形态学特征, 可将它们人为划分5个阶段, 即营养生长期(0~31 d)、抽穗前期(32~65 d)、抽穗期(66~90 d)、成熟期(91~135 d)和完全成熟期(136~160 d)。由表2、表3和图1可见, 在Cd处理条件下, OsMT1、 OsDCT1、 OsLCD、OsPCs、OsPCR6、OsPCR9和 OsLCT1基因表达水平的差异不显著( P>0.05), 而 OsPCR1基因表达水平的差异显著( P<0.05)。说明, OsPCR1基因可能参与水稻体内Cd积累的调控。
| 表2 5 μmol L-1 CdCl2处理24 h后, 秀水110和秀水11叶片中Cd积累相关基因的表达水平(营养生长期)(平均值± SE) Table 2 Expression level of Cd-accumulation-associated genes in leaves of Xiushui 110 and Xiushui 11 after 24 h of 5 μmol L-1CdCl2treatment (vegetative stage) (mean ± SE) |
| 表3 5 μmol L-1 CdCl2处理60 d后秀水110和秀水11叶片中Cd积累相关基因的表达水平(抽穗期)(平均值± SE) Table 3 Expression level of Cd-accumulation-associated genes in leaves of Xiushui 110 and Xiushui 11 after 60 days of 5 μmol L-1 CdCl2treatment (pre-heading stage) (mean ± SE) |
为研究 OsPCR1基因表达调控的分子调控机制, 通过RGAP和谷类小RNA数据库(Cereal Small RNA Database, CSRDB)[ 40]分析发现, smRNA92891 (siRNA: 5'-GCGGTCCCAGTACGGCCTGCAGG-3') 在 OsPCR1基因的外显子2对应的mRNA存在潜在靶向位点。由图1和图2可见, 在Cd处理条件下, 营养生长期、抽穗前期和完全成熟期水稻叶片中, siRNA的表达水平与各自相对应时期 OsPCR1基因的表达水平呈负相关(siRNA的低水平表达对应 OsPCR1基因的高水平表达)。并且由图3可见, 在Cd处理条件下, 秀水110叶片中, OsPCR1基因表达水平的时间变化曲线和该siRNA表达水平的时间变
化曲线是完全相反的( OsPCR1基因表达水平的上升和该siRNA表达水平的下降同时发生)。图3提示, 在Cd处理条件下的秀水11中, OsPCR1基因表达水平的时间变化曲线和该siRNA表达水平的时间变化曲线虽然不是完全相反的, 但是呈现出互补性。表明该siRNA可能参与调控 OsPCR1基因的表达。
在对照条件下, 除了抽穗期, 水稻叶片中该siRNA的表达水平和 OsPCR1基因的表达水平没有呈现出负相关性之外, 其他时期均呈现出负相关性。并且由图3可见, 对照条件下的两个水稻品种叶片中, OsPCR1基因和该siRNA表达水平的时间变
化曲线也呈现出互补性。由于siRNA的积累和mRNA的降解都是量变的过程, 所以对照条件下的抽穗期siRNA和 OsPCR1mRNA没有呈现出负相关性可能是由于该siRNA积累速率较慢(图1-C、图2-C、图3-C和D)。Cd处理条件下的抽穗期和成熟期siRNA和 OsPCR1mRNA没有呈现出负相关性可能是由于转录水平的调控(比如DNA甲基化)(图1-C, D和图2-C, D)。在Cd处理条件下, 由于两个水稻品种中siRNA的水平在这两个时期无显著的差异(图2-C, D), 所以siRNA在转录后水平对 OsPCR1的调控不明显, 而转录水平的调控(比如DNA甲基化)对
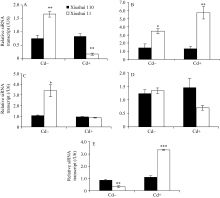 | 图2 不同生长发育期的秀水110和秀水11叶片中siRNA基因的表达水平利用qRT-PCR技术检测水稻品种倒数第2叶和第3叶总siRNA表达水平。数据以U6核小RNA为内参进行标准化定量, 以各自秀水110的样本为参照(定义为1), 以平均值±标准误表示(mean ± SE) ( n = 3~4)。A~E对应时期, 标注及缩写同图1。Fig. 2 siRNA expression in leaves of Xiushui 110 and Xiushui 11 at five developmental stages of riceqRT-PCR was performed to detect the expression of the siRNA in the penultimate and antepenultimate leaves of the two cultivars. Relative expression of siRNA in the leaves of Xiushui 11 was normalized to that in the leaves of Xiushui 110, which is defined as 1. U6 small nuclear RNA was used for normalization. The data are presented as mean ± SE. ( n = 3 and n = 4). The stages in individual figure, symbols and abbreviation are the same as those given in Fig. 1. |
基因的表达调控影响更大。表明该siRNA的积累很可能在转录后降解了 OsPCR1基因mRNA从而参与调控 OsPCR1基因的表达。
为检测水稻中 OsPCR1基因表达的差异是否由于DNA甲基化的调节, 分析了秀水110和秀水11叶片中 OsPCR1基因外显子2甲基化水平在水稻5个不同生长发育期中的变化情况。由图1和图4可见, 在Cd处理条件下, DNA甲基化水平与各自相对应时期水稻叶片中 OsPCR1基因的表达水平呈负相关(DNA的高水平甲基化对应 OsPCR1基因的低水平表达)。并且由图3-A和B可见, 在Cd处理条件下, 秀水11和秀水110叶片中, DNA甲基化水平的时间变化曲线和 OsPCR1基因表达水平的时间变化曲线基本呈现出互补性。表明甲基化可能参与调控
OsPCR1基因的表达。
而在对照条件下, 不同生长发育期水稻叶片中 OsPCR1基因外显子2甲基化水平(图4)与各自相对应时期的 OsPCR1基因的表达水平(图1)没有负相关性。且对照条件下水稻叶片中, DNA甲基化水平的时间变化曲线和 OsPCR1基因表达水平的时间变化曲线没有互补性(图3)。由于siRNA对基因的表达调控发生在转录后水平, 而DNA甲基化对基因表达的调控发生在转录水平; 并且在对照条件下, 各生长时期2个水稻品种间该siRNA的量存在显著差异(图2-A, B, C, E)。所以这可能是由于siRNA对 OsPCR1基因在转录后水平表达的调控掩盖了DNA甲基化对 OsPCR1基因在转录水平表达的调控。由图3-A可见, 当该siRNA丰度降低到低水平时, DNA甲基化的程度逐渐缓和并且不再发生大的变化。而由图3-A、B和C可见, 当该siRNA积累时,
 | 图3 不同生长发育期的秀水110和秀水11叶片中 OsPCR1基因、siRNA的表达水平和DNA甲基化水平提取5个不同生长发育期秀水110和秀水11倒数第2和第3叶基因组DNA和总RNA。利用McrBC-qRT-PCR检测 OsPCR1外显子2甲基化水平, qRT-PCR检测 OsPCR1基因表达水平和siRNA表达水平。相对 OsPCR1外显子2甲基化水平的数据校准到等量未经酶切的基因组DNA。 OsPCR1基因的表达水平校准到 OsUBQ5。siRNA的表达水平校准到U6小核仁RNA。所有数据以各自营养生长期的样本为参照, 以平均值±标准误表示(mean ± SE) ( n=3~8)。 OsPCR1: OsPCR1相对表达水平; siRNA: siRNA相对表达水平; Unme: OsPCR1外显子2相对未甲基化水平。标注及缩写同图1。Fig. 3 OsPCR1 expression level, siRNA expression level, and OsPCR1 exon 2 methylation level in leaves of (A) Xiushui 110 and (B) Xiushui 11 at five developmental stages of riceGenomic DNA and total RNA were isolated from leaves previously described in Fig. 1 legend. OsPCR1 exon 2 methylation, OsPCR1 and siRNA expressions were determined by the methods of McrBC-qRT-PCR and qRT-PCR, respectively. Relative unmethylated OsPCR1 exon 2 level, OsPCR1 mRNA and siRNA expressions were normalized to equal amounts of undigested DNA samples, OsUBQ5 and U6 small nuclear RNA, respectively and relative to the vegetative samples. The data are presented as mean ± SE ( n = 3 to n = 8). OsPCR1: relative OsPCR1expression level; siRNA: relative siRNA level; Unme: relative unmethylated OsPCR1 exon 2 level. Symbols and abbreviation are the same as those given in Fig. 1. |
 | 图4 不同生长发育期的秀水110和秀水11叶片中 OsPCR1基因外显子2甲基化水平提取5个不同生长发育期秀水110和秀水11倒数第2和第3叶基因组DNA。利用特异性的引物和McrBC-qRT-PCR技术检测 OsPCR1基因外显子2甲基化水平。数据校准到等量未经酶切的DNA样本, 以各自秀水110的样本为参照(定义为1), 以平均值±标准误表示(mean ± SE) ( n = 3~4)。A~E对应时期, 标注及缩写同图1。Fig. 4 OsPCR1exon 2 methylation in leaves of Xiushui 110 and Xiushui 11 at five developmental stages of riceGenomic DNA was isolated from leaves described in Fig. 1 legend. OsPCR1exon 2 methylation was determined by the methods of McrBC-qRT-PCR with specific primers. The data were normalized to equal amount of undigested DNA samples and relative to Xiushui 110 samples, respectively. The data are presented as means ± SE ( n = 3 and n = 4). The stages in individual figure, symbols and abbreviation are the same as those given in Fig. 1. |
DNA甲基化的程度会发生大的变化。说明siRNA的积累和DNA甲基化的动态变化是同时发生的。该siRNA的积累可能直接或间接地影响 OsPCR1基因外显子2区域的DNA甲基化水平; OsPCR1基因外显子2甲基化和该siRNA可能同时参与调控 OsPCR1基因的表达。
许多Cd积累相关基因参与调节植物体内Cd的转移和积累[ 5, 6, 7, 8, 9]。本文研究表明, 2个水稻品种中 OsMT1、 OsDCT1、 OsLCD、OsPCs、OsPCR6、OsPCR9和 OsLCT1基因表达水平的差异不显著。暗示报道过的 OsMT1、 OsDCT1、 OsLCD、OsPCs和 OsLCT1基因可能没有直接导致秀水11和秀水110叶片中Cd积累的差异。
PCR蛋白参与拟南芥体内Cd从地下部到地上部的运输[ 9, 12]。水稻中PCR家族成员的功能尚不明确,研究表明, 两个水稻品种间 OsPCR1基因表达水平差异显著, 而O sPCR6和 OsPCR9基因的表达水平差异不显著。进一步的研究表明, 处于不同生长发育期的2个水稻品种叶片中, OsPCR1基因的表达水平始终呈现出显著的差异。提示 PCR基因家族成员 OsPCR6和 OsPCR9与水稻叶片中Cd积累的差异可能没有直接关系。而 OsPCR1基因的表达差异可能与水稻叶片中Cd积累的差异相关。这为以后研究该基因在水稻体内Cd积累过程中的功能提供了理论依据。
植物体内的siRNAs参与发育过程和逆境胁迫反映[ 13, 14, 15, 16, 17]。siRNAs通过与靶基因的特异性结合而达到降解靶基因mRNA的作用[ 18]。本实验表明, 各生长发育期水稻叶片中siRNA的表达水平和 OsPCR1基因表达水平此消彼长。提示该siRNA可能与AGO蛋白结合参与形成RISC沉默复合体, 剪切 OsPCR1基因的mRNA, 从而在转录后水平上沉默 OsPCR1基因的表达。说明siRNA在水稻体内Cd积累的过程中发挥重要作用。
DNA甲基化是一种表观遗传修饰标记, 在植物的生长发育和逆境胁迫响应以及转座子的沉默和基因的表达调控中都有重要作用[ 21, 22, 23, 24, 25, 26, 27]。在基因启动子区域的DNA甲基化抑制基因的表达[ 41], 而基因编码区域DNA甲基化修饰的功能尚不明确[ 29, 32, 42]。为此, 我们分析了水稻叶片中 OsPCR1基因外显子2甲基化水平在各生长发育期的变化情况, 及DNA甲基化水平和 OsPCR1基因表达水平的相关性。结果表明, 在Cd胁迫下, 水稻叶片中 OsPCR1外显子2甲基化水平与 OsPCR1基因的表达水平此消彼长。提示 OsPCR1外显子2甲基化可能参与调控 OsPCR1基因的表达。基因编码区域DNA甲基化修饰和基因转录的延伸有关[ 43, 44, 45]。与此模型一致, 研究结果暗示 OsPCR1基因编码区域DNA甲基化修饰可能参与调控 OsPCR1基因转录的延伸。本试验表明在Cd胁迫和对照条件下的生长发育过程中, 水稻叶片中 OsPCR1基因外显子2甲基化水平均出现动态变化。而动态的基因外显子区域DNA甲基化可通过影响基因转录物的选择性剪接来调控基因的表达[ 46, 47]。本研究结果同样表明 OsPCR1外显子甲基化修饰也可能通过参与转录产物的剪切来调控 OsPCR1基因的表达。说明在水稻体内Cd积累过程中, DNA甲基化修饰发挥重要作用。
在Cd高积累和Cd低积累水稻品种叶片中Cd积累相关基因的表达水平差异不显著, 但 OsPCR1基因表达水平呈现显著差异。说明 OsPCR1基因可能参与调控水稻体内Cd的积累。这为后续研究该基因的功能提供了理论依据。水稻叶片中与 OsPCR1基因外显子区域匹配的siRNA的丰度和 OsPCR1基因的表达水平负相关, 且在Cd处理条件下的水稻叶片中 OsPCR1基因外显子区域DNA甲基化水平和该基因的表达水平负相关。说明在水稻体内Cd积累过程中, 该siRNA和 OsPCR1基因外显子区域DNA甲基化可能参与 OsPCR1基因表达的调控。
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
|
| [45] |
|
| [46] |
|
| [47] |
|